The script analyses the functional differences between glycolytic enzymes using principal component analysis (PCA) and hierarchical clustering. It generates some very nice heat maps.
Here's one of the plots, produced with the pheatmap package
 |
| Analysing enzymes in terms of bond cleavage |
SCRIPT
### Title: The chemical logic of glycolysis
### Author: Sergio Martínez Cuesta, EMBL-EBI, Hinxton, UK
### Aim: To visualise chemical similarities between enzymes reactions in glycolysis using latent variable models (PCA and CA) and clustering techniques
## Data input: load bond change data into R
data <- read.csv("http://science2therapy.com/data/bc.csv", header=TRUE, row.names=1, sep="\t", check.names=FALSE)
# bc.csv contains the frequency counts of chemical bond changes of the 10 glycolytic reactions as obtained from EC-BLAST
# header=TRUE and row.names=1 allow the first row and column to be considered the column and row names, sep="\t" indicates that bc.csv is tab-separated and check.names=FALSE disables the check of variable names
## Explore data
data
class(data)
dim(data)
colnames(data) # types of bond changes (formed and cleaved)
rownames(data) # enzyme ids, see data/glycolysis_enzymes.csv
## Barplots
# Which glycolytic enzyme catalyses more bond changes?
freqs_bc<-sort(rowSums(data), decreasing=TRUE)
freqs_bc
barplot(freqs_bc, col="white", ylab="Frequency of bond changes", xlab="Enzyme name")
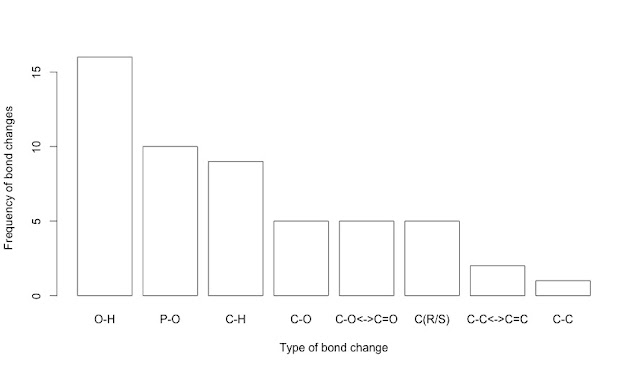
# Which bond changes are more common in glycolysis?
freqs_enz<-sort(colSums(data), decreasing=TRUE)
freqs_enz
barplot(freqs_enz, col="white", ylab="Frequency of bond changes", xlab="Type of bond change")
# Average number of bond changes by glycolytic reaction
mean(rowSums(data))
## Principal component analysis (PCA)
fit <- princomp(data)
summary(fit)
biplot(fit, main="PCA")
loadings(fit)
fit$scores
# Additional packages: FactoMineR
install.packages("FactoMineR") # this only needs to be done once
library(FactoMineR)
fit <- PCA(data, scale.unit = FALSE)
## Correspondence analysis (CA)
install.packages("ca") # this only needs to be done once
library(ca)
fit <- ca(data)
summary(fit)
plot(fit)
# using library(FactoMineR)
# fit<-CA(data)
## Calculate bond change distances between glycolytic reactions
?dist
d <- dist(data, method = "manhattan")
## Hierarchical clustering
fit <- hclust(d, method="ward")
plot(fit, main="Hierarchical clustering")
groups <- cutree(fit, k=4)
rect.hclust(fit, k=4, border="red")
## Heatmaps
heatmap(as.matrix(data))
heatmap(as.matrix(d))
## Pretty heatmaps
install.packages("pheatmap") # this only needs to be done once
library(pheatmap)
pheatmap(data,
fontsize=13,
cellwidth = 15,
cellheight = 15,
color = colorRampPalette(c("white", "red"))(4),
clustering_distance_rows=d,
legend_breaks=c(0,1,2,3))
## How to save pdf images:
pdf("mygraph.pdf")
pheatmap(data)
dev.off()
# References:
# 1- Quick-R: http://www.statmethods.net/
# 2- Crawley, M. J. (2007). The R Book. Chichester, UK: John Wiley & Sons, Ltd. doi:10.1002/9780470515075
# 3- Rahman, S. A., Martínez Cuesta, S., Furnham, N., Holliday, G. L., & Thornton, J. M. (2014). EC-BLAST: a tool to automatically search and compare enzyme reactions. Nature methods, 11(2), 171–174. doi:10.1038/nmeth.2803. url: http://www.ebi.ac.uk/thornton-srv/software/rbl/
# 4- Husson, F., Lê, S., and Pagès, J. (2011) Exploratory multivariate analysis by example using R, [online] http://books.google.com/books?hl=en&lr=&id=EDnNBQAAQBAJ&oi=fnd&pg=PP1&dq=Exploratory+Multivariate+Analysis+by+Example+Using+R&ots=I4bnnmgPJw&sig=ENW39pnO00fOOSO_F01cQ_Iv-UA (Accessed February 6, 2015).
# 5- Keller, M. A., Turchyn, A. V., & Ralser, M. (2014). Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Molecular systems biology, 10, 725. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/24771084








No comments:
Post a Comment
Comments and suggestions are welcome.