Today, I published a piece in The Biochemist about data visualisation with R. I enjoyed writing the piece and it includes some of the code and blog posts that I have written here.
The code I used to create the figures has all been uploaded on Github. To reproduce the figures you can cut and paste the code from there into a script window on R-Studio.
Here is a list of pages with code for the figures.
Figure 1 - R allows reproducible data visualisations.
- Figure 1A - My first published R data visualization – a cluster dendrogram from a proteomic data set. First published in 2014 and reproduced in 2021 (Alsagaby et al, 2014, https://doi.org/10.1021/pr5002803).
- Figure 1B - A violin plot showing neutralizing antibody titres against five SARS-CoV-2 strains reproduced from shared data and code from Wall et al (2021, https://doi.org/10.1016/S0140-6736(21)01290-3).
- Figure 1C - Phylogenetic tree of human kinase domains inspired by visualization by Manning et al (2002, https://doi.org/10.1126/science.1075762).
- Figure 2A - A volcano plot.
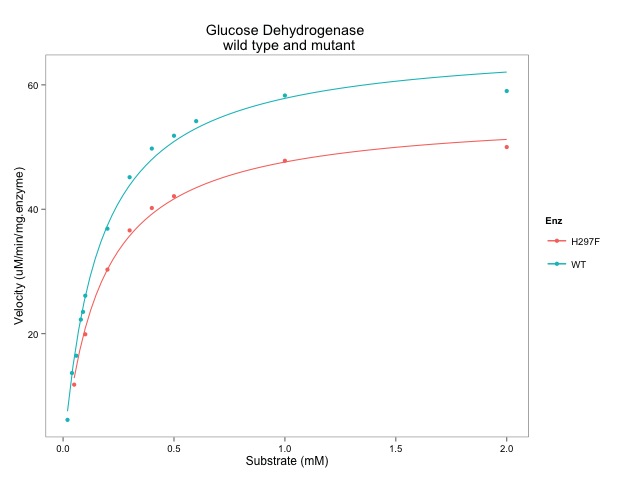
- Figure 2B - A drug dose–response curve and LD50 calculation.
- Figure 2C - A simple flow cytometry plot.
- Figure 3A - A volcanic activity time line inspired by @ijeamaka_a
- Figure 3B - Illustrating the importance of numbers in password strength. Across a range of password types, inclusion of numbers increases password strength.
- Figure 3C (Github only) - Showing the proportion of female culprits in Scooby Doo shows from 1960s to 2020s.
- Figure 4A - The Hexsticker for the Bioconductor package drawProteins. (b)
- Figure 4B (Github only) - Schematic of SARS-CoV S1 protein variants. Protein and variant information from Uniprot (https://www.uniprot.org/uniprot/P0DTC2).