As part of my job, I'm required to check the
TravelHealthPro website which contains information about the recommended vaccinations when visiting other countries. For example, for a visit to
Namibia, TravelHealthPro advises that most visitors have vaccinations to:
- Hepatitis A
- Tetanus
- Typhoid
When the
Cardiff R User Group decided to discuss and work on web scraping, I decided to scrape the
TravelHealthPro web site and prepare some maps with selected recommended vaccinations.
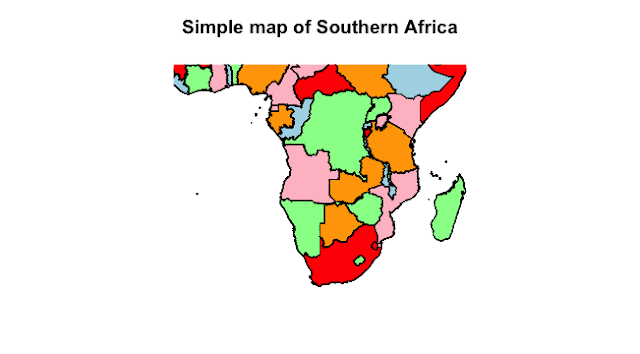
Here are some of the world maps, I've prepared:
The script involved some webscraping, some data reformatting and then the visualisations. Here is all the R script involved in making these maps. It was originally prepared as separate scripts to keep it easier for me to understand but I have combined them all here in parts.
For just the mapping part, it is possible to start halfway with a download from Github of the scraped data (as of April 26th, 2017). To do that start at Part 3.
Feedback welcome as always.
# START SCRIPT
library("tools") # uses the toTitleCase() function
library("rvest") # tidyverse webscraping package
library("curl")
library("RCurl") # for downloading data from github
library("rworldmap") # for the maps
## PART 1: scrape the list of countries from TravelPro Home page
# scrape the page.
# this is the URL
url <- c("http://travelhealthpro.org.uk/countries")
# readLines() is a base R function which allows reading html from pages
data <- readLines(url)
print(paste(url, "has just been scraped"))
# identify where the proteins names are on the scraped data
country_numbers <- grep("travelhealthpro.org.uk/country/", data)
# 317 long... sounds like a good length
# function for removing html tags
# based on http://stackoverflow.com/questions/17227294/removing-html-tags-from-a-string-in-r
extractCountry <- function(htmlString) {
htmlString <- gsub("<.*?>", "", htmlString)
htmlString <- gsub("\t", "", htmlString)
return(htmlString)
}
extractCountry(data[country_numbers[4]])
# function for extracting the url for each country
extractCountryUrl <- function(htmlString) {
htmlString <- gsub("\\t<li><a href=", "", htmlString)
htmlString <- gsub("</a></li>", "", htmlString)
htmlString <- gsub("\"", "", htmlString)
htmlString <- gsub(">.*", "", htmlString)
return(htmlString)
}
# test the function
extractCountryUrl(data[country_numbers[7]])
# loop through the character vector and create a vector of countries
# and a vectors of URLs
countryList <- NULL
countryUrls <- NULL
for(i in 1:length(country_numbers)){
reqd_url <- extractCountryUrl(data[country_numbers[i]])
country <- extractCountry(data[country_numbers[i]])
countryList <- c(countryList, country)
countryUrls <- c(countryUrls, reqd_url)
}
# I have a list of countries and a list of URLs
countryList # up to 276 looks ok - Zimbabwe should be last
# truncate at Zimbabwe
zimb <- grep("Zimbabwe", countryList)
countryList <- countryList[1:zimb]
countryUrls <- countryUrls[1:zimb]
## PART 2: with list of countries, extract vaccination info...
# I am attempting to scrape vaccinations recommendations
# from a Travel Health Pro website on a particular country.
# the example here is travel health pro page about afghanistan
# this is the page
# http://travelhealthpro.org.uk/country/1/afghanistan#Vaccine_recommendations
# scraping the webpage is all in this function
extractVacs <- function(x){
# scrape the page
web_content <- read_html(curl(x, handle = new_handle("useragent" = "Chrome")))
# handle is required as extra data and curl package is required too
print(paste(x, "has just been scraped"))
# extracting the data using pipes
vac_list_most <- web_content %>%
html_node(".accordion") %>% # Note this is html_node - first one (Most Travellers)
html_nodes(".accordion-item") %>%
html_node("p") %>%
html_text(trim = FALSE)
vac_list_some <- web_content %>%
html_nodes(".accordion") %>% # Note this is html_nodes - all vaccinations!
html_nodes(".accordion-item") %>%
html_node("p") %>%
html_text(trim = FALSE)
# using gsub to remove the spaces and the line brake symbol
vac_list_most <- gsub("\n", "", vac_list_most)
vac_list_most <- gsub(" ", "", vac_list_most)
vac_list_some <- gsub("\n", "", vac_list_some)
vac_list_some <- gsub(" ", "", vac_list_some)
#substract vac_list_most from vac_list_some
vac_list_some <- setdiff(vac_list_some, vac_list_most)
countryName <- gsub("http://travelhealthpro.org.uk/country/", "", x)
countryName <- gsub("[[:digit:]]+", "", countryName)
countryName <- gsub("/", "", countryName)
countryName <- toTitleCase(countryName)
# make the list with country name and the two vaccinations lists...
vac_list <- list(country = countryName, vac_most = vac_list_most, vac_some = vac_list_some)
# this works and returns a list.
return(vac_list)
}
# test the code
vac_list_example <- extractVacs(countryUrls[2])
vac_list_example
# works
# for demo purposes - just scrape four
output_demo <- lapply(countryUrls[7:10], extractVacs)
# to scrape all the countries, remove the hash tag and run the whole thing
# output <- lapply(countryUrls, extractVacs)
# it's a good plan to save this file so that it doesn't have to be scraped again
# save(output, file = "vaccinationList_scraped20170426")
# and save the country list
# save(countryList, file = "countryList_scraped20170426")
## PART 3: If you don't want to scrape the data - download it...
link <- "https://raw.githubusercontent.com/brennanpincardiff/RforBiochemists/master/data/countryList_scraped20170426.rda"
download.file(url=link, destfile="file.rda", mode="wb")
countryList <- readRDS("file.rda")
link <- "https://raw.githubusercontent.com/brennanpincardiff/RforBiochemists/master/data/vaccinationList_scraped20170426.rda"
download.file(url=link, destfile="file.rda", mode="wb")
output <- readRDS("file.rda")
# add "Ivory Coast" to the country List for future binding into rworldmap
countryList <- c(countryList, "Ivory Coast")
# add a new element to list with Ivory Coast
ivory <- output[grep("Ivory", output)]
ivory[[1]]$country <- c("Ivory Coast")
ivory[[1]]
output <- c(output, ivory[1]) # just one square bracket N.B.
# so we have a list of vaccinations and countries that is 277 long
# have a list of 277 with the vaccinations in...
# and looks good.
# next step - turn these two objects into something to do a visualiation.
## PART 4: Clean up vaccinations list
# get the whole unique list of vaccinations/drugs is not trivial
# inconsistent labelling on the site
vac_req_mostTrav <- unique(unlist(lapply(output, '[[', 2)))
# white space issue for Hepatitis A
vac_req_someTrav <- unique(unlist(lapply(output, '[[', 3)))
# trailing white space and capitalisation issues for TBE
# "Tick-Borne Encephalitis (TBE)"
# "Tick-Borne Encephalitis (TBE) "
# "Tick-borne encephalitis (TBE)"
# "Tick-borne encephalitis (TBE) "
# apply toTitleCase and some gsubs in each element in output
for(i in 1:length(output)){
output[[i]] <- lapply(output[[i]], toTitleCase)
output[[i]]$country <- gsub("-", " ", output[[i]]$country)
output[[i]]$vac_most <- gsub("Hepatitis a", "Hepatitis A", output[[i]]$vac_most)
output[[i]]$vac_some <- gsub("Hepatitis a", "Hepatitis A", output[[i]]$vac_some)
output[[i]]$vac_some <- gsub("Tick-Borne Encephalitis (TBE) ",
"Tick-Borne Encephalitis (TBE)",
output[[i]]$vac_some)
}
# toTitleCase will change Hepatitis A to Hepatitis a so change it back...
## PART 5: Pull out countries with vaccinations we're interested in...
# want a data frame with country in one column
# then I want a column entitled Hep A, Polio etc...
vac_df <- data.frame(countryList, stringsAsFactors = FALSE)
colnames(vac_df) <- c("country")
# go through each element in the list (lapply)
# use a function to see if the vaccination I want is required...
# example: Polio
polioStatus <- lapply(1:length(output), function(x){"Polio" %in% output[[x]]$vac_most})
vac_df$polio <- unlist(polioStatus)
vac_df$polio_val <- as.numeric(vac_df$polio)
# Yellow Fever
yellFevStatus <- lapply(1:length(output), function(x){"Yellow Fever" %in% output[[x]]$vac_most})
vac_df$yellFev <- unlist(yellFevStatus)
vac_df$yellFev_val <- as.numeric(vac_df$yellFev)
# Tick-Borne Encephalitis (TBE)
tbeStatus <- lapply(1:length(output), function(x){"Tick-Borne Encephalitis (TBE)" %in% output[[x]]$vac_some})
vac_df$tbe <- unlist(tbeStatus)
vac_df$tbe_val <- as.numeric(vac_df$tbe)
## PART 6: join the data into the countries in rworldmap package
#join to a coarse resolution map
spdf <- joinCountryData2Map(vac_df, joinCode="NAME", nameJoinColumn="country")
# 211 codes from your data successfully matched countries in the map
# 66 codes from your data failed to match with a country code in the map
# 32 codes from the map weren't represented in your data
# where polio vaccination is recommended to most travellers
mapCountryData(spdf, nameColumnToPlot="polio_val",
catMethod="fixedWidth",
addLegend = FALSE,
mapTitle = "Polio Vaccination Recommended")
text(0,-90, "Source: http://travelhealthpro.org.uk/")
# where yellow fever vaccination is recommended to most travellers
mapCountryData(spdf, nameColumnToPlot="yellFev_val",
catMethod="fixedWidth",
addLegend = FALSE,
mapTitle = "Yellow Fever Vaccination Recommended")
text(0,-90, "Source: http://travelhealthpro.org.uk/")
# where Tick-Borne Encephalitis vaccination is recommended to some travellers
mapCountryData(spdf, nameColumnToPlot="tbe_val",
catMethod="fixedWidth",
addLegend = FALSE,
mapTitle = "Tick-Borne Encephalitis \nVaccination Recommended (some travellers)")
text(0,-90, "Source: http://travelhealthpro.org.uk/")
# zoom on Africa for Yellow Fever data
mapCountryData(spdf, nameColumnToPlot="yellFev_val",
catMethod="fixedWidth",
mapRegion = "Africa",
addLegend = FALSE,
mapTitle = "Yellow Fever Vaccination Recommended")
text(10,-35, "Source: http://travelhealthpro.org.uk/")
## END SCRIPT
Useful Resources and further reading