Using R to generate visualizations of biological data
(Paul Brennan & Dean Hammond)Tuesday, 8th March 2016 at VizBi 2016 Conference
Summary
The aim of the tutorial is to provide an opportunity for participants to learn how to use R, the open source statistical language, to illustrate their data. Through the use of R-Studio, a graphical user interface, participants can see how their data is manipulated, how data can be selected and manipulated and how this can be used to generate figures for their thesis or for publication.
Topics
Learning objectives
Process
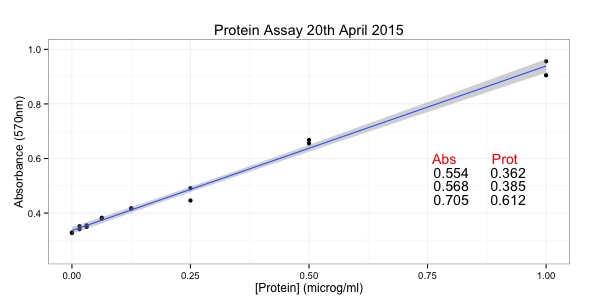
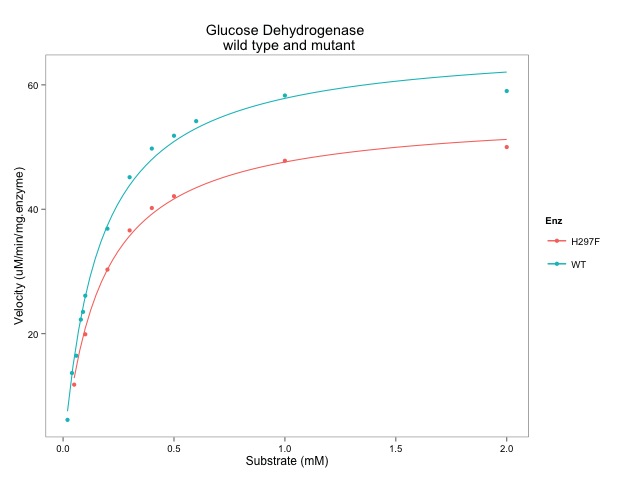
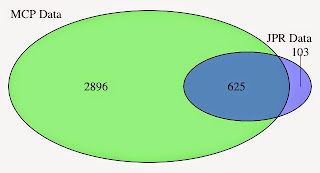
The course will be structured around R scripts which will generate visualisations of biological data. Participants will be led through the scripts and encouraged to run the scripts on their own machines. Changes that occur in R-Studio will be discussed and described. The scripts will be available prior to and after the tutorial. One hour will be spent on each of the three learning objectives. It will be an interactive session with help from at least one additional tutor. Approximately six scripts will be illustrated over the course of the tutorial. At least one of the tutors will be available during the VizBi Conference for further discussion.
For further information visit the R for Biochemists blog
or contact a tutor: Dr Paul Brennan (BrennanP@cardiff.ac.uk); @brennanpcardiff

No comments:
Post a Comment
Comments and suggestions are welcome.