I'm preparing some teaching materials for another Biochemical Society R training event with the draft title of R for Biochemists 201. Some more advanced material based on feedback for participants of R for Biochemists 101.
In preparation, I've been looking at published proteomics data. I've come across a nice paper by a group in the Barts Cancer Institute in London. The paper is entitled "Proteomic and genomic integration identifies kinase and differentiation determinants of kinase inhibitor sensitivity in leukemia cells". It was published in the journal Leukaemia.
I visited their lab once many years ago and I have heard the senior author, Pedro Cutillas, talk. It is very interesting work, I think.
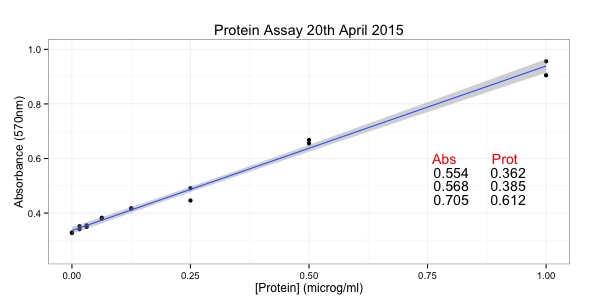
They have made their data available so I've spend some time writing a script that makes one part of Figure 1a - a nice box and whisker plot.
Here is the plot:
START
## data import
library(readxl)
library(ggplot2)
link <- "https://static-content.springer.com/esm/art%3A10.1038%2Fs41375-018-0032-1/MediaObjects/41375_2018_32_MOESM2_ESM.xlsx"
download.file(link, "temp_data")
data <- read_excel("temp_data", skip=2)
# skip = 2 stops the first two rows being part of the file
# the next row is used as titles of the columns
# skip needs to be determined by looking at the data
# remove bottom two rows as only 36 patients
data <- data[1:36,]
# using the geom_boxplot() function, we can draw our graph
ggplot(data = data,
aes(FAB, log10(`MEKi (trametinib)...13`), colour = FAB)) +
geom_boxplot(na.rm = TRUE)
# we can make it look a bit more like the plot in the paper using geom_jitter()
plot <- ggplot(data = data,
aes(FAB, log10(`MEKi (trametinib)...13`), colour = FAB)) +
geom_boxplot(na.rm = TRUE) +
geom_jitter(width=0.15, na.rm = TRUE) +
theme_bw() +
labs( y = "Log10(EC50)nM",
title = "Sensitivity of AML patients samples to MEK inhibitor",
subtitle = "Casado et al (2018) Leukaemia 32:1818–1822
doi:10.1038/s41375-018-0032-1") +
theme(legend.position="none")
plot
# to print a high resolution of this the tiff() function can be used.
tiff("plot.tiff", height = 12, width = 17, units = 'cm', compression = "lzw", res = 300)
plot
dev.off()
END
Some resources: