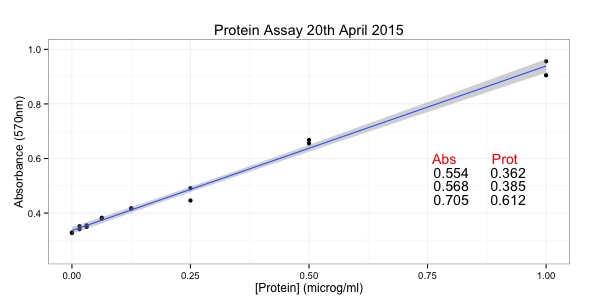
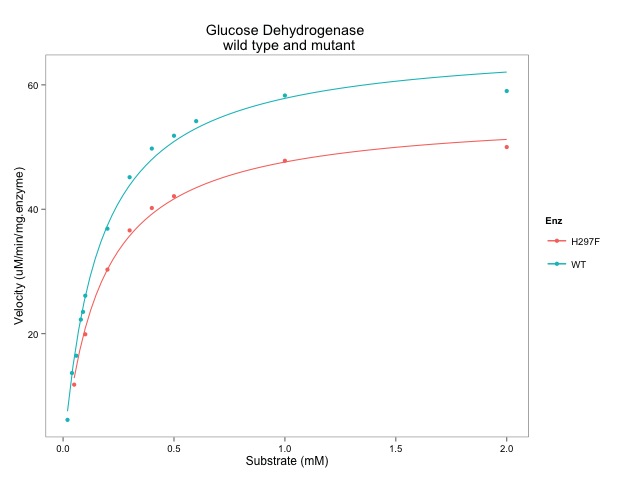
Could you point me in the direction of help to make a plot with different axes? For example for a character series “fractions” make one plot with two sets of data, one plotted against the right and one against the left y axes where the two y axes could be something like absorbance and activity. Thanks!
This inspired me to create this visualisation:
If you more information, leave a comment below.
Here is the script:
START
#https://stackoverflow.com/questions/43942002/geom-line-with-different-y-axis-scale
library(magrittr)
library(tidyr)
sample_data <- data.frame(fracs = rep(1:20),
Absorbance = c(0.1, 0.1, 0.1, 0.2, 0.4, 0.1, 0.1, 0.5, 1.0, 0.1,
0.1, 0.1, 0.5, 0.1, 0.1, 0.2, 0.3, 0.5, 1.0, 0.1),
Activity = c(32, 33, 35, 150, 287, 39, 35, 40, 42, 32,
30, 31, 32, 45, 57, 32, 33, 40, 41, 40))
# reformat the data into long format to allow plotting
data <- sample_data %>% gather(y, value, Absorbance:Activity)
data$fracs <- as.integer(data$fracs)
# trial plot for Activity (enzyme activity)
ggplot(data, aes(fracs, value, colour=y)) +
geom_line(data=data %>% filter(y=="Activity"))
# trial plot for Absorbance
ggplot(data, aes(fracs, value, colour=y)) +
geom_line(data=data %>% filter(y=="Absorbance"))
# plot the two lines together
ggplot(data, aes(fracs, value, colour=y)) +
geom_line(data=data %>% filter(y=="Absorbance")) +
geom_line(data=data %>% filter(y=="Activity"))
# but different values makes Absorbance line flat.
# change the scale for second line
max(sample_data$Absorbance)
max(sample_data$Activity)
scaleby = max(sample_data$Activity)/max(sample_data$Absorbance)
ggplot(data, aes(colour=y)) +
geom_line(data=data %>% filter(y=="Activity"),
aes(x = fracs, y = value)) +
geom_line(data=data %>% filter(y=="Absorbance"),
aes(x = fracs, y=value*scaleby))
# this looks better...
# putting it together with labels etc...
p <- ggplot() +
geom_line(data=data %>% filter(y=="Activity"),
aes(x = fracs, y = value, colour=y)) +
geom_line(data=data %>% filter(y=="Absorbance"),
aes(x = fracs, y=value*scaleby, colour=y))
# limit for y axis
p <- p + ylim(0,300)
# adding our second scale on the right hand side of the plot
p <- p + scale_y_continuous(sec.axis = sec_axis(~./scaleby,
name="Absorbance (570nM)"))
# label the x and y axis...
p <- p + ylab("Enzyme activity (units)")
p <- p + xlab("Chromatography fractions")
# This is salt chromatography so adding a line showing
# the NaCl concentration.
salt_conc <- data.frame(fracs = rep(1:20),
conc = c(c(10), seq(from = 10, to = 250, by= 13)))
# salt curve labels
salt_label <- data.frame(x = c(1,20),
y = c(10, 250),
text = c("[10nM]\nNaCl", "[250nM]\nNaCl"))
# check out the line
ggplot(salt_conc, aes(fracs, conc)) +
geom_line() +
geom_text(data = salt_label,
aes(x = x,
y = y,
label = text))
# linetype=2 gives a dashed line
p <- p + geom_line(data = salt_conc, aes(fracs, conc),
linetype = 2)
# add some labels to show the starting and ending concentrations
p <- p + geom_text(data = salt_label,
aes(x = x,
y = y,
label = text),
size = 2.5)
p <- p + theme_bw()
p <- p + ggtitle("Chromatography plot")
p <- p + theme(legend.position="top", legend.direction="horizontal")
p <- p + theme(legend.title=element_blank())
p
END








No comments:
Post a Comment
Comments and suggestions are welcome.