I have chosen a paper from Molecular and Cellular Proteomics which uses aptamers to detect multiple proteins in exosomes and cells.
The data was generated by colleagues in the School of Medicine at Cardiff University including the first author - Dr Jason Webber, a Prostate Cancer UK funded Research Fellow and senior author, Dr Aled Clayton, a Senior Lecturer in Cancer & Genetics at Velindre Hospital. Dr Tim Stone was key to the data analysis. The protein detection method is from a company called SomaLogic.
The first step was downloading the file from the Molecular and Cellular Proteomics website. I used the download.file() function. This saves the file into your current directory. This was opened using the readxl package (by Hadley Wickham) using the read_excel() function.
I drew some graphs as I explored and manipulated the data. These are interspersed with the script below. The visualisations included boxplots and a cluster diagram.
I wanted to draw a volcano plot which expresses the fold change against the significant of the change (p-value). I couldn't do that with the data supplied so I had to reverse the transformation and calculate the fold change again.
Here is the volcano plot:
 |
| Comparison of changes in exosomes compared cells. Proteins over-expressed in exosomes are on the right. Proteins over-expressed in cells are on the left. |
The plot indicates that there are more proteins over-expressed in cells (on the left) compared to exosomes (on the right).
Here is the script with some other plots:
START
# pull down a file from the internet, do some analysis and draw a graph...
# choose Jason Webber's MCP Paper...
# http://www.mcponline.org/content/13/4/1050.full
# supplementary data is here: http://www.mcponline.org/content/suppl/2014/02/06/M113.032136.DC1/mcp.M113.032136-5.xlsx
# install if necessary:
# install.packages(c("ggplot2", "readxl"))
library(ggplot2)
library(readxl)
# this is the link to the data
link <- "http://www.mcponline.org/content/suppl/2014/02/06/M113.032136.DC1/mcp.M113.032136-6.xlsx"
# the download.file() function downloads and saves the file with the name given
download.file(url=link,destfile="file.xlsx", mode="wb")
# then we can open the file and extract the data using the read_excel() function.
data<- read_excel("file.xlsx")
View(data)
# plot the data - always an important first step!
boxplot(data[2:7],
las =2, # las = 2 turns the text around to show sample names
ylab = expression(bold("expression")),
main="Boxplot of expression data")
 |
| Two types of sample - exosomes (n=3) and cells (n=3). |
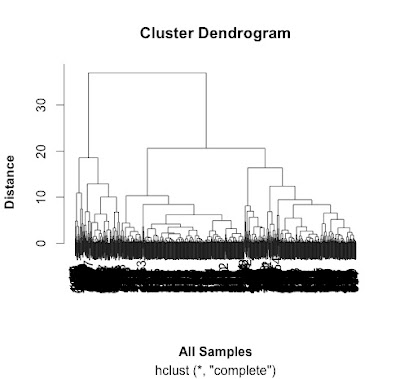
# do a cluster analysis to quality control the different groups
# convert to matrix first
data.m <- as.matrix(data[2:7])
dim(data.m) # gives the dimensions of the matrix
# ans: 762 6
# calculate the distances using the dist() function.
# various methods are possible - default is Euclidean.
distances <- dist(data.m)
summary(distances) # have a look at the object
# make the cluster dendrogram object using the hclust() function
hc <- hclust(distances)
# plot the cluster diagram
# some interesting groups in the data
plot(hc,
xlab =expression(bold("All Samples")),
ylab = expression(bold("Distance")))
# ah, not what I intended.
# it clustered the proteins NOT the samples.
# transpose the data and try again...
data.m.t <- t(data.m)
dim(data.m.t)
# ans = 6 762 - so that has worked.
# repeat cluter analysis
# calculate the distances using the dist() function.
# various methods are possible - default is Euclidean.
distances <- dist(data.m.t)
summary(distances)
# make the cluster dendrogram object using the hclust() function
hc <- hclust(distances)
# plot the cluster diagram
# some interesting groups in the data
plot(hc,
xlab =expression(bold("All Samples")),
ylab = expression(bold("Distance")))
# replicates cluster together well.
# the adjusted P value are in a column entiteld: BH - P.value
# this is a little awkward so rename this and Fold Change column:
colnames(data)[9] <- "P.Value"
colnames(data)[10] <- "Fold.Change"
plot(data$P.Value ~ data$Fold.Change)
# this works but we would like to turn it into a volcano plot
# with log2 at the bottom and -p-value.
# we can't log fold changes as half of them are negative numbers
# we need to re-calculate the raw data
# reverse the log2 transformation.
# mean the values & calculate fold change in decimal format (no +/-)
data$exo1 <- 2^data$exoRFU1
data$exo2 <- 2^data$exoRFU2
data$exo3 <- 2^data$exoRFU3
data$cell1 <- 2^data$cellRFU1
data$cell2 <- 2^data$cellRFU2
data$cell3 <- 2^data$cellRFU3
# calculate means of the replicates
data$exoMean <- rowMeans(data[,13:15])
data$cellMean <- rowMeans(data[,16:18])
# always good to visualise the data:
plot(log2(data$exoMean)~log2(data$cellMean))
# calculate fold change exo/cell
data$FoldChange <- data$exoMean/data$cellMean
plot(log2(data$FoldChange)) # transform for plotting
# make column in data.frame with transformed data
data$Log2.Fold.Change <- log2(data$FoldChange)
## Identify the genes that have a p-value < 0.05
data$threshold = as.factor(data$P.Value < 0.05)
## Construct the volcano plot object using ggplot
g <- ggplot(data=data,
aes(x=Log2.Fold.Change, y =-log10(P.Value),
colour=threshold)) +
geom_point(alpha=0.4, size=1.75) +
xlim(c(-6, 6)) +
xlab("log2 fold change") + ylab("-log10 p-value") +
theme_bw() +
theme(legend.position="none") +
ggtitle("Volcano Plot comparing protein expression in exosomes vs cells ") # add a title
g # show the plot